RAPIDEC® CARBA NP
A simple and efficient test for carbapenemase detection in Enterobacteriaceae and Pseudomonas aeruginosa.
New FDA 510(k) cleared test detects carbapenemase-producing bacteria grown in culture. The test provides results in 30 minutes to 2 hours, making it the easy way to detect carbapemase producers and to help you improve patient management and control healthcare-associated infections (HAI).
- Ready to use
- Easy to perform
- Results when you need them
The carbapenemase global burden
Rising antimicrobial drug resistance is one of the most urgent challenges facing the field of healthcare today. Carbapenem antibiotics are last-resort drugs used to treat bacterial infections. However, strains of carbapenem-resistant bacteria, particularly Enterobacteriaceae, are rapidly emerging.
There are 2 types of bacterial resistance to carbapenems. If resistance results from impermeability or efflux, it is not transmissible to other bacteria and there is no need to isolate patients to prevent the spread of infection. If the resistance is caused by carbapenemase enzyme production, it is transmissible and preventative action is necessary. Infections caused by carbapenemase producers are therefore very difficult to treat, have a high mortality rate and can easily spread between humans via contact, food and water – leading to greater risk of outbreaks of healthcare-associated infections (HAI). In Antibiotic Resistance Threats 2013, the CDC classes carbapenem resistance in its most serious category – urgent threat – meaning it is “an immediate public health threat that requires urgent and aggressive action.”1
Detection of resistance to carbapenems, particularly when due to transmissible carbapenemase-producing bacteria, is critical to properly orient antibiotic therapy and implement appropriate isolation measures. Most methods for detection have long time-to-results and/or are complicated and costly. What’s more, some tests lack sensitivity and specificity, making results less reliable.
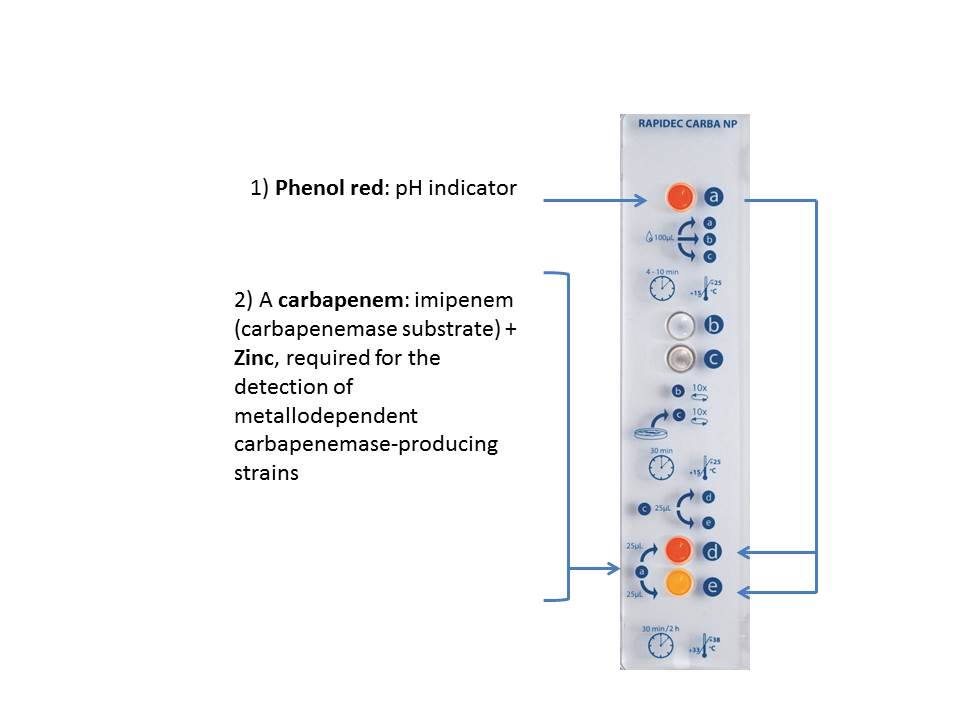
The Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing recommends per M100-S27 (January 2017) that laboratories using Enterobacteriaceae MIC breakpoints for carbapenems as described in M100-S20 (January 2010) perform the Modified Hodge with suspected carbapenem-resistant organisms. The procedure has a high amount of hands-on time, is difficult to both perform and interpret, and has been shown to have false positive results.2 CLSI also recommends the Carba NP test, based upon the work of Nordmann and Poirel.3,4 The Carba NP test is based upon the bacterial carbapenemase enzyme hydrolyzing imipenem in the presence of zinc. The hydrolysis of imipenem causes a change in pH which causes phenol red in the medium to change color from red to yellow. To run the Carba NP test the laboratory must gather the necessary reagents to construct the test. Most laboratories do not have the time or capacity to perform the complex procedure and are looking for a ready-made solution that provides same days results for carbapenemase detection.
RAPIDEC CARBA NP: A simple and efficient solution
The new FDA 510(k) cleared RAPIDEC CARBA NP offers high medical value as a cost-effective, all-in-one solution providing results in 30 minutes to 2 hours. The RAPIDEC CARBA NP is a phenotypic (colorimetric) in vitro diagnostic test for the qualitative detection of carbapenemase enzymes in Enterobacteriaceae and Pseudomonas aeruginosa. With its sensitivity of 99.6% and specificity of 97.4% for the routine subculture procedure, and 98.5% and 97.3% in the short subculture procedure, the test can contribute to better patient management to help control the spread of HAI.
RAPIDEC CARBA NP provides same day carbapenemase detection results in an all-inclusive test kit.
- Adaptable to any lab
- Easy to use
- Cost-effective
- Same-day results
- Manage infection risk: reliable, sensitive and specific
Compared to other methods, RAPIDEC CARBA NP is…
…easy to perform
…less costly (versus molecular methods)
What makes RAPIDEC CARBA NP different is how it works. It was developed in collaboration with international experts P. Nordmann and L. Poirel, based on the principle they described along with colleague L. Dortet.3,4 The RAPIDEC CARBA NP test detects carbapenem hydrolysis by carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa grown in culture. Hydrolysis acidifies the medium which results in a color change of the pH indicator – indicating the presence of a carbapenemase.
RAPIDEC CARBA NP detects (without distinction) all 3 types of produced carbapenemases:
- Class A: KPC is the main representative
- Class B: Metallo ß-Lactamases (MBL). NDM, VIM and IMP
- Class D: OXA-48
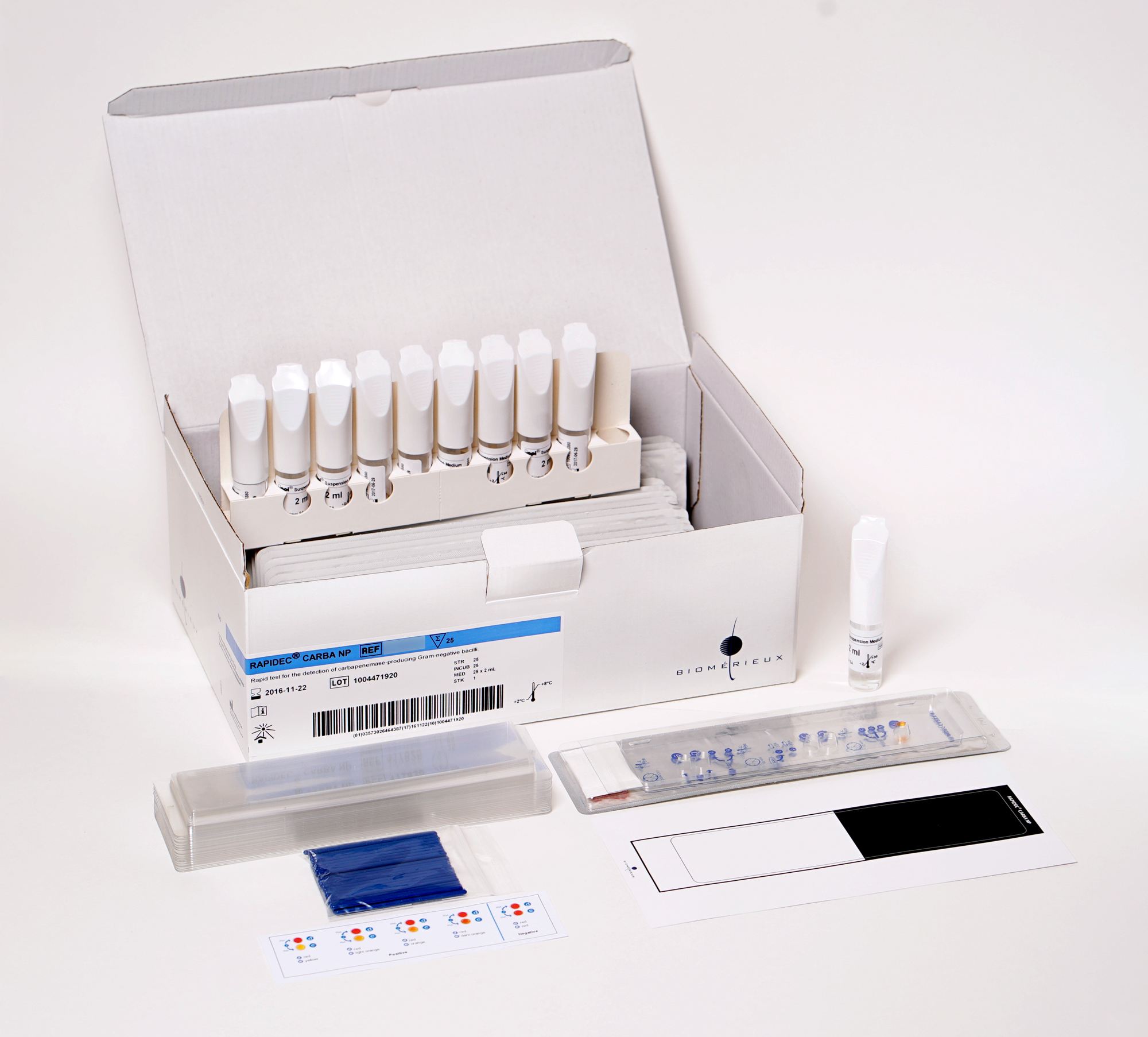
Ready-to-use kit
The ready-to-use RAPIDEC CARBA NP kit contains all you need to conduct the test in a few easy steps.
- After bacterial lysis, which enables the extraction of the enzyme, the lysate is added to a detection solution which contains:
- After incubation, readings are performed by visually comparing a control well, starting at 30 minutes, and after no more than 2 hours.
Each kit contains:
- Test strips
- Ampules of API® Suspension Medium, 2 mL
- Incubation lids
- 1 pack of stirring sticks
- Two-colored (black and white) reading support card
The package insert and reading guide are available to download from www.mybioMerieux.com
- Antibiotic Resistance Threats in the United States, CDC 2013
- Cecilia G, Carvalhaes C, Picao R, Nicoletti A, Xavier D, Gales A. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother. 2010;65:249-251. jac.oxfordjournals.org/content/65/2/249.full.pdf+html
- Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerging Infectious Diseases, www.cdc.gov/eid, 2012;8(9):1503-1507.
- Dortet L, Poirel L, Nordmann P. Rapid identification of carbapenemase types using a biochemical test in Enterobacteriaceae and Pseudomonas. Antimicrob Agents Chemother. 2012;56(12):6437-6440.
RAPIDEC CARBA NP Kit Reference Number
Kit for 10 tests (ref. 417824)
- 10 test strips
- 10 ampules of API® Suspension Medium, 2 mL
- 10 incubation lids
- 1 sachet of stirring sticks
- 1 two-colored (black and white) support
- 1 package insert provided in the kit or downloadable from www.mybioMerieux.com
- Reading guide downloadable from www.mybioMerieux.com
Please contact your local bioMérieux representative for product availability.
Performance
CLINICAL PERFORMANCE
In the RAPIDEC CARBA NP clinical study, a total of 457 strains consisted of 394 Enterobacteriaceae and 63 Pseudomonas aeruginosa were evaluated across 5 sites using both the routine and the short subculture procedures.
Carbapenemase determination was determined by a composite reference method composed of 3 tests: carbapenem MIC (Imipenem, Meropenem, Ertapenem and/or Doripenem), CLSI® Carba NP and carbapenemase genetic markers by Polymerase Chain Reaction (PCR).
Results from those three different tests were used to determine the carbapenemase positive/negative status of an isolate, with the final composite reference result based on agreement of at least two of the three tests. Agreement between RAPIDEC CARBA NP and composite reference method was assessed. When a RAPIDEC CARBA NP result was not in agreement with the composite reference result, it was further evaluated. A negative RAPIDEC CARBA NP result was considered as a false negative when the composite reference result was determined to be positive, indicating a false non-carbapenemase producer. A positive RAPIDEC CARBA NP result was considered as a false positive when the composite reference result was determined to be negative, indicating a false carbapenemase producer. The comparative performance is shown in Table 1 below.
Table 1: Comparative Performance of RAPIDEC CARBA NP (Enterobacteriaceae + Pseudomonas aeruginosa)
| Incubation | Total # | Agreement # | Agreement % | Negative # | Positive # | False Positivea # (0%) | False Negativeb #(0%) |
| Routine Subculture | 457 | 451 | 98.7 | 192 | 265 | 5 (2.6) | 1 (0.4) |
| Short Subculture | 449 | 440 | 98.0 | 188 | 261 | 5 (2.7) | 4 (1.5) |
a False positive for carbapenemase; RAPIDEC CARBA NP positive result for a non-carbapenemase producing Enterobacteriaceae or P. aeruginosa.
b False negative for carbapenemase; RAPIDEC CARBA NP negative result for a carbapenemase producing Enterobacteriaceae or P. aeruginosa.
The RAPIDEC CARBA NP test was evaluated with Enterobacteriaceae and Pseudomonas aeruginosa colonial growth on sheep blood agar medium incubated for 18-24 hours (routine subculture) and at least 4 hours (short subculture). An initial RAPIDEC CARBA NP reading was performed at 30 minutes and if negative a final reading was performed at a total of 2 hours of 33-38°C incubation.
The routine subculture/incubation procedure included 457 samples in which 306 were clinical samples provided by the investigational sites and 151 were well-characterized challenge samples. It included 394 Enterobacteriaceae and 63 Pseudomonas aeruginosa with carbapenemase enzymes described below:
- 265 samples were positive for carbapenemase enzyme: 147 KPC, 52 NDM, 26 VIM, 17 IMP and 23 OXA-48.
- 192 samples were carbapenemase enzyme negative.
- The short subculture/incubation procedure included 449 samples in which 300 were clinical samples provided by the investigational sites and 149 were well-characterized challenge samples. It included 392 Enterobacteriaceae and 57 Pseudomonas aeruginosa with carbapenemase enzymes described below:
- 261 samples were positive for carbapenemase enzymes: 146 KPC, 52 NDM, 23 VIM, 17 IMP and 23 OXA-48.
- 188 samples were carbapenemase enzyme negative.
The performance of the RAPIDEC CARBA NP test with target carbapenemases and non-target carbapenemases is demonstrated in Table 2.
| Carbapenemase determination by composite reference method | RAPIDEC CARBA NP performance | ||||
| Routine Subculture/Incubation | Short subculture/incubation | ||||
| N | Agreement (%) | N* | Agreement (%) | ||
| Enterobacteriaceae | 383/394 | 98.5% | 383/392 |
97.7% |
|
| Pseudomonas aeruginosa | 63/63 | 100% | 57/57 | 100% | |
| All samples | 451/457 | 98.7% | 440/449 | 98.0% | |
| Positive samples | 264/265a | 99.6% | 257/261c | 98.5% | |
| Enterobacteriaceae |
KPC NDM VIM IMP OXA-48 |
143/144a 51/51 15/15 12/12 23/23 |
99.3% 100% 100% 100% 100% |
142/143c 50/51c 14/15c 12/12 22/23c
|
99.3% 98.0% 93.3% 100% 95.7% |
| Total | 244/245 | 99.6% | 240/244 | 98.4% | |
| Pseudomonas aeruginosa |
KPC NDM IM IMP |
3/3 1/1 11/11 5/5 |
100% 100% 100% 100% |
3/3 1/1 8/8 5/5 |
100% 100% 100% 100% |
| Total | 20/20 | 100% | 17/17 | 100% | |
| Negative Samples | 187/192b | 97.4% | 183/188d | 97.3% | |
|
Enterobacteriacae
|
|
144/149b
|
96.6%
|
143/148d
|
96.6% |
| Pseudomonas aeruginosa | 43/43 | 100% | 40/40 | 100% | |
* Insufficient growth/biomass for six P. aeruginosa and two K.pneumoniae in the short subculture procedure, they were three VIM producing P. aeruginosa, and one KPC producing K. pneumoniae; four negative samples including three P. aeruginosa and one K. pneumoniae.
a Routine subculture false negative (false non-carbapenemase producer) rate was 0.4% (1/265) for claimed carbapenemases; the false negative was KPC-producing Enterobacteriaceae
b Routine subculture false positive (false carbapenemase producer) rate was 2.6% (5/192) for P. aeruginosa and Enterobacteriaceae; the five false positives were from Enterobactericeae
c Short subculture false negative (false non-carbapenemase producer) rate was 1.5% (4/257) for claimed carbapenemases; one false negative each for KPC, NDM, VIM, and OXA-48 from Enterobacteriaceae
d Short subculture false positive (false carbapenemase producer) rate was 2.7% (5/188); the five false positives were from Enterobacteriaceae
LIMITATIONS OF THE METHOD
- The performance of the RAPIDEC CARBA NP assay for detection of carbapenemases enzymes encoded by genetic markers other than KPC, NDM, OXA-48, VIM and/or IMP has not been established. In addition, RAPIDEC CARBA NP results may be influenced by the local epidemiology regarding genetic markers of resistance, i.e., depending on the local distribution/prevalence of different carbapenemase genetic markers, and more false negative results may occur. Conduct alternative testing if negative results are obtained and carbapenemase enzyme production is suspected based on local epidemiology.
- RAPIDEC CARBA NP testing should be used as an adjunct to other laboratory test(s) such as antimicrobial susceptibility testing.
- The performance of the RAPIDEC CARBA NP test with bacteria other than Enterobacteriaceae and Pseudomonas aeruginosa has not been evaluated. Organism identification and elevated carbapenem MICs should be determined prior to testing on the RAPIDEC CARBA NP.
- Proteus species, Providencia species, Morganella species may have elevated imipenem MICs due to intrinsic resistance mechanisms. Pseudomonas aeruginosa has been shown to exhibit resistance to ertapenem due to intrinsic resistance mechanisms.
- The detection of OXA variants other than OXA-48 has not been evaluated sufficiently in the study.
- Hypermucoid colonies may lead to false positive or false negative results and should not be tested by the RAPIDEC CARBA NP test. A hypermucoid colony tends to stretch itself to form a continuous viscous filament > 5 mm in length when picked up from an agar plate using a bacteriology loop/needle (6).
- Agar media containing pH indicator for colony color differentiation (e.g., Bromocresol Purple, MacConkey, Cysteine Lactose Electrolyte-Deficient, etc.) are not compatible with the RAPIDEC CARBA NP and require subculturing growth/biomass on a sheep blood agar for testing.
- The performance of RAPIDEC CARBA NP has been evaluated for subculturing growth on 5% sheep blood agar incubated for 18-24 hours (Routine procedure) and 4-5 hours (Short Incubation procedure) only. The performance with other culture media has not been evaluated and is therefore unknown.
- The performance of the RAPIDEC CARBA NP test when testing Enterobacteriaceae and Pseudomonas aeruginosa containing OXA-181, OXA-232, SME, GIM, SPM, and IMI carbapenemase enzymes has not been established due to the low number of positive isolates available using the Composite Reference Method.